In-Vivo MRI Tangles Marker
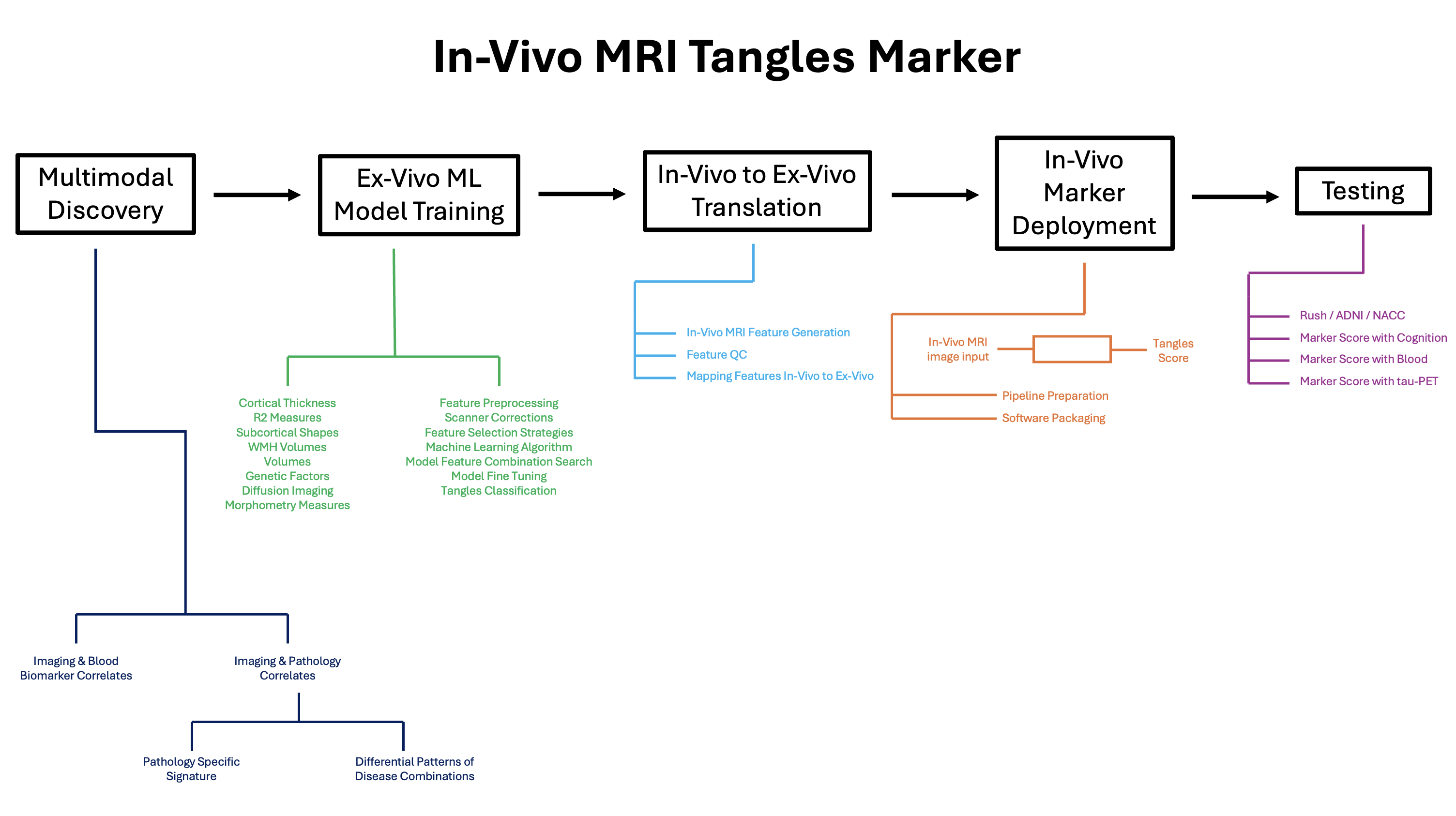
What this figure shows
This visual summary illustrates my end-to-end workflow for building an in-vivo MRI marker of neurofibrillary tangles. The central idea is to discover pathology-anchored imaging signatures in an autopsy cohort, train and refine models on ex-vivo MRI (where pathology is known), and then translate those signatures to in-vivo MRI so the marker can be used in living cohorts.
Pipeline (left → right)
- Multimodal discovery: identify candidate imaging features and biomarker/pathology correlates to define hypothesis-driven signatures. [Volume and Shape patterns] [AD and LATE patterns]
- Ex-vivo ML model training: train models using ex-vivo MRI features (e.g., morphometry/DTI/R2/WMH and related measures), with careful preprocessing and feature selection to predict tangles burden.
- In-vivo → ex-vivo translation: generate and QC in-vivo features and map them onto the ex-vivo feature space to enable consistent scoring across modalities.
- Marker deployment: package the pipeline so an in-vivo MRI input yields a tangles score.
- Testing: validate the marker in external and/or independent datasets (e.g., Rush / ADNI / NACC) and evaluate associations with cognition, blood biomarkers, and tau-PET.
Why it matters
A scalable, MRI-based tangles score can help stratify participants, track neurodegeneration, and support clinical trial enrichment—especially when PET or invasive measures are limited.
Resources
ISMRM 2022 · Abstract
AAIC 2022 · Abstract
For related work from our lab, see ARTS [Paper] [Software] (in-vivo MRI marker for arteriolosclerosis)] and MARBLE [Paper] (in-vivo MRI marker for LATE).
